PEAKS AB 3.5https://crackeddownload.com/product/peaks-ab-3-5/
PEAKS AB 3.5 Software New FeaturesPEAKS AB automates protein de novo antibody sequencing using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) datasets from orthogonal enzyme digests. From confident de novo sequence tags, the full protein sequence is assembled using a weighted de Brujin graph1. In peaks AB 3.5, the software package is equipped with additional tools to enhance sequencing accuracy, PTM characterisation and result presentation including: - Retention time prediction for isobaric differentiation.
- Improved intact mass deconvolution algorithm.
- lle/Leu differentiation by the signature w-ions included in sequence validation function.
- Added support for in-depth O-glycan analysis.
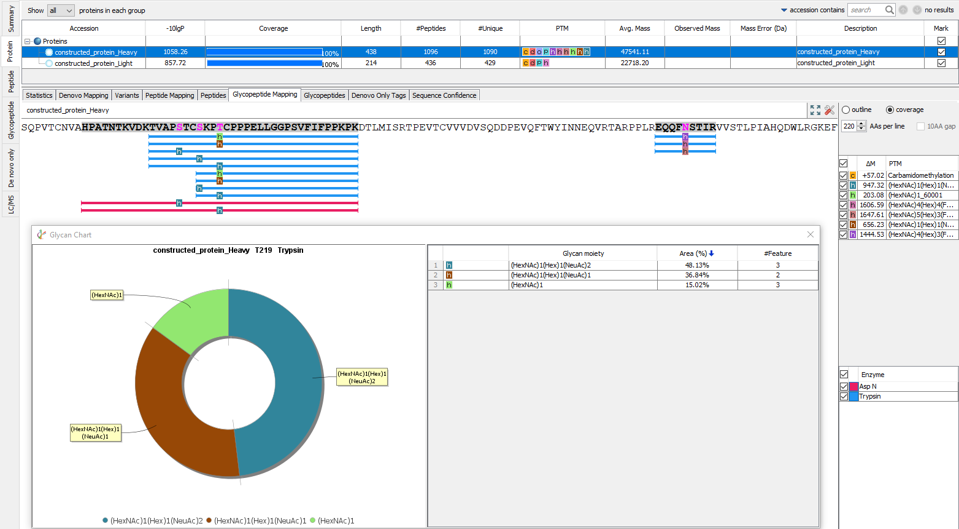
- Merged glycopeptides into peptide mapping view.
- Support for timsTOF data
- Improved UI and report.
In-Depth Glycan ProfilingIn our previous PEAKS AB build we introduced N-linked glycan profiling. In addition to this, PEAKS AB 3.5 now supports identification and profiling of O-linked glycans. This tool performs in-depth glycan profiling for both N- and O-linked sites identified in the heavy and/or light chains of the antibody. In addition to accurate glycopeptides mapping to the assembled antibody sequence, enzyme-based glycan profiling displays the composition and relative abundance of each glycan at a selected glycosylation site. Glycan composition and structure annotation are provided within each glycopeptide spectrum and are based on glycan fragment ions that match to an O-linked or N-linked glycan databases. Accurate localisation of the glycan at each site is achieved by identifying fragment ions of glycan moieties associated with the peptide backbone.
Retention Time Prediction to Differentiate Isobaric Residues
| 
